Table of Contents
External Links for netCDF
Maintainer
Introduction
In the climate computing netCDF is often used as a container for two types of data: the grid and the measurements. Typically, a grid is represented as a set of points, whereas each of them represents a position on the earth surface. Many models use pairs of longitude and latitude values for this purpose, but there are also more complex ones, that include height, vertices, topography, and so on. To each such a grid point we can assign a set of different values, like temperature, air pressure, or insolation. We also need to take into the account, that these values can change over time.
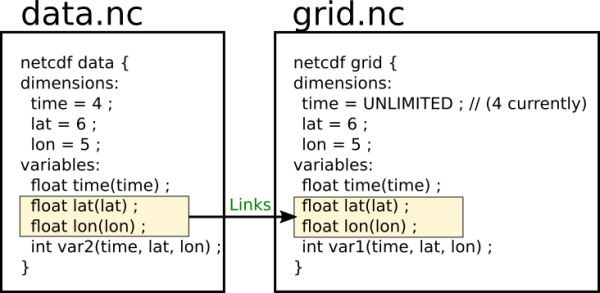
In netCDF a simple grid can be constructed by two dimensions, e.g. longitude and latitude. Changes over time can be represented by another dimension, e.g. time. These dimensions can be assigned to a variable which contains our measurements. As a result we get a variable, where each value is assigned to longitude, latitude and time step. In a netCDF we can put several such variables and connect them to the same dimensions. But as soon we start using several files we get duplication of the grid, because each file must contain one. Depending on the size and complexity of the grid, it can consume a significant amount of disk space. Therefore we need to find a way to reuse the grid.
An obvious solution to this problem is to store the grid in one file and create links to the grid in the other files. But the current netCDF version is missing such a feature. We decided to extend netCDF and wrote a patch.
Detailed Description
Behind the API of the current implementation of netCDF-4 interface hides the HDF5 library, that has a wide range of useful features, which we can use to implement the link functionality in netCDF. We decided to use HDF5 Virtual Datasets (VDS), a feature that was introduced in the HDF5-1.10.0. VDS is powerful feature of HDF5 and it provides more functionality that is needed for our purpose. Our patch takes a simple usage of it. It looks at the dimensionality of the source dataset and create a virtual dataset with the same dimensionality in the target file. Infinite dimensions are not supported yet.
Emulations of netCDF dimensions in HDF5 is realized by HDF5 datasets and relies on a heavy usage of HDF5 attributes. More precisely, for each dimension netCDF creates a dataset stored and attaches a set of different attributes. The dataset can store dimension labels and the attributes contains meta information about the dimensions, e.g. index, name, attached variables. When using virtual datasets for creating links to datasets the attributes are not created automatically. This work must be done manually. The attributes “CLASS”, “NAME”, and “REFERENCE_LIST”, are can be easily created by the HDF5 scale interface. The attribute “_Netcdf4Dimid” is a pure netCDF component and is created by the HDF5 attribute interface.
Although, HDF5 allows to create virtual datasets even if the target datasets don't exist, we couldn't make our patch to work in this way. To work properly our patch requires information from the source file, like dimensionality, datatype. This implies, that the target file and the valid datasets must exist at runtime.
When for some reason, after the links are created, the target file becomes inaccessiable (e.g. deleted, renamed, unreadable, …) and the target dataset is not available the links will be filled with default values. In our case it is the value 0.
Our patch introduces a new function:
int nc_def_dim_external(int ncid, const int dimncid, const char *name, int *idp)
| Name | Type | Description |
|---|---|---|
| ncid | in | File id. Links will be created here. |
| dimncid | in | File id, where dimensions are located. |
| name | in | dimension name |
| idp | out | dimension id |
Measurements
| Model | Resolution | Grid size | Data size |
|---|---|---|---|
| HD(CP)2 | 2323968 cells | 2.8GB | 100GB |
| HD(CP)2 | 7616120 cells | 9.1GB | 340GB |
| HD(CP)2 | 22282304 cells | 27GB | 1000GB |
Internal structure of datafiles with high resolution:
$ h5ls 3d_fine_day_DOM03_ML_20130502T141200Z.nc
bnds Dataset {2}
clc Dataset {1/Inf, 150, 22282304}
cli Dataset {1/Inf, 150, 22282304}
clw Dataset {1/Inf, 150, 22282304}
height Dataset {150}
height_2 Dataset {151}
height_bnds Dataset {150, 2}
hus Dataset {1/Inf, 150, 22282304}
ncells Dataset {22282304}
ninact Dataset {1/Inf, 150, 22282304}
pres Dataset {1/Inf, 150, 22282304}
qg Dataset {1/Inf, 150, 22282304}
qh Dataset {1/Inf, 150, 22282304}
qnc Dataset {1/Inf, 150, 22282304}
qng Dataset {1/Inf, 150, 22282304}
qnh Dataset {1/Inf, 150, 22282304}
qni Dataset {1/Inf, 150, 22282304}
qnr Dataset {1/Inf, 150, 22282304}
qns Dataset {1/Inf, 150, 22282304}
qr Dataset {1/Inf, 150, 22282304}
qs Dataset {1/Inf, 150, 22282304}
ta Dataset {1/Inf, 150, 22282304}
time Dataset {1/Inf}
tkvh Dataset {1/Inf, 151, 22282304}
ua Dataset {1/Inf, 150, 22282304}
va Dataset {1/Inf, 150, 22282304}
vertices Dataset {3}
wa Dataset {1/Inf, 151, 22282304}
Download
netCDF patch
- Depends on HDF5-1.10.0 or higher
- Adds external dimensions to netCDF-library
- no support for unlimited dimensions
- no support for external variables
HDF5 patch
- Workaround for the “Assertion `ret == NC_NOERR' failed.” issue.
Installation
- HDF5
- Download and extract HDF5-1.10.0-patch1
- Download and extract the patch to the HDF5 directory
- Change to the HDF5 directory
- Apply the patch
git apply hdf5-1.10.0-patch1.patch - Configure, make and install HDF5 in the usual way
- netCDF
- Download and extract netCDF-c-4.4.1-rc2
- Download and extract the patch to the netCDF directory
- Change to the netCDF directory
- Apply the patch
git apply netcdf-c-4.4.1-rc2.patch - Configure, make and install netCDF in the usual way
Usage
int nlat, dimid; int grid_ncid, data_ncid; const char* gridfile = "grid.nc"; const char* datafile = "data.nc"; nc_open(gridfile, NC_NOWRITE, &grid_ncid); nc_create(datafile, NC_NETCDF4, &data_ncid); nc_def_dim_external(data_ncid, grid_ncid, "lat", &dimid);
Example
Step 1: Creating a netCDF file
In the first step a netCDF4 file “grid.nc” is created. It contains labeled dimensions “lat”, “lon” and “time”, and a variable “var1”. The output of ncdump shows the header of the file.
Download: mkncfile.c
Output of ncdump:
$ ncdump -h grid.nc
netcdf grid {
dimensions:
time = UNLIMITED ; // (4 currently)
lat = 6 ;
lon = 5 ;
variables:
float time(time) ;
float lat(lat) ;
float lon(lon) ;
int var1(time, lat, lon) ;
}
Output h5dump:
$ h5dump -A -p grid.nc
HDF5 "grid.nc" {
GROUP "/" {
ATTRIBUTE "_NCProperties" {
DATATYPE H5T_STRING {
STRSIZE 8192;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_UTF8;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "version=1|netcdflibversion=4.4.1-rc2|hdf5libversion=1.10.0"
}
}
DATASET "lat" {
DATATYPE H5T_IEEE_F32LE
DATASPACE SIMPLE { ( 6 ) / ( 6 ) }
STORAGE_LAYOUT {
CONTIGUOUS
SIZE 24
OFFSET 17098
}
FILTERS {
NONE
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE 9.96921e+36
}
ALLOCATION_TIME {
H5D_ALLOC_TIME_LATE
}
ATTRIBUTE "CLASS" {
DATATYPE H5T_STRING {
STRSIZE 16;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "DIMENSION_SCALE"
}
}
ATTRIBUTE "NAME" {
DATATYPE H5T_STRING {
STRSIZE 4;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "lat"
}
}
ATTRIBUTE "REFERENCE_LIST" {
DATATYPE H5T_COMPOUND {
H5T_REFERENCE { H5T_STD_REF_OBJECT } "dataset";
H5T_STD_I32LE "dimension";
}
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): {
DATASET 11799 /var1 ,
1
}
}
}
ATTRIBUTE "_Netcdf4Dimid" {
DATATYPE H5T_STD_I32LE
DATASPACE SCALAR
DATA {
(0): 1
}
}
}
DATASET "lon" {
DATATYPE H5T_IEEE_F32LE
DATASPACE SIMPLE { ( 5 ) / ( 5 ) }
STORAGE_LAYOUT {
CONTIGUOUS
SIZE 20
OFFSET 17122
}
FILTERS {
NONE
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE 9.96921e+36
}
ALLOCATION_TIME {
H5D_ALLOC_TIME_LATE
}
ATTRIBUTE "CLASS" {
DATATYPE H5T_STRING {
STRSIZE 16;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "DIMENSION_SCALE"
}
}
ATTRIBUTE "NAME" {
DATATYPE H5T_STRING {
STRSIZE 4;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "lon"
}
}
ATTRIBUTE "REFERENCE_LIST" {
DATATYPE H5T_COMPOUND {
H5T_REFERENCE { H5T_STD_REF_OBJECT } "dataset";
H5T_STD_I32LE "dimension";
}
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): {
DATASET 11799 /var1 ,
2
}
}
}
ATTRIBUTE "_Netcdf4Dimid" {
DATATYPE H5T_STD_I32LE
DATASPACE SCALAR
DATA {
(0): 2
}
}
}
DATASET "time" {
DATATYPE H5T_IEEE_F32LE
DATASPACE SIMPLE { ( 4 ) / ( H5S_UNLIMITED ) }
STORAGE_LAYOUT {
CHUNKED ( 1024 )
SIZE 4096
}
FILTERS {
NONE
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE 9.96921e+36
}
ALLOCATION_TIME {
H5D_ALLOC_TIME_INCR
}
ATTRIBUTE "CLASS" {
DATATYPE H5T_STRING {
STRSIZE 16;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "DIMENSION_SCALE"
}
}
ATTRIBUTE "NAME" {
DATATYPE H5T_STRING {
STRSIZE 5;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "time"
}
}
ATTRIBUTE "REFERENCE_LIST" {
DATATYPE H5T_COMPOUND {
H5T_REFERENCE { H5T_STD_REF_OBJECT } "dataset";
H5T_STD_I32LE "dimension";
}
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): {
DATASET 11799 /var1 ,
0
}
}
}
ATTRIBUTE "_Netcdf4Dimid" {
DATATYPE H5T_STD_I32LE
DATASPACE SCALAR
DATA {
(0): 0
}
}
}
DATASET "var1" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 4, 6, 5 ) / ( H5S_UNLIMITED, 6, 5 ) }
STORAGE_LAYOUT {
CHUNKED ( 1, 6, 5 )
SIZE 480
}
FILTERS {
NONE
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE -2147483647
}
ALLOCATION_TIME {
H5D_ALLOC_TIME_INCR
}
ATTRIBUTE "DIMENSION_LIST" {
DATATYPE H5T_VLEN { H5T_REFERENCE { H5T_STD_REF_OBJECT }}
DATASPACE SIMPLE { ( 3 ) / ( 3 ) }
DATA {
(0): (DATASET 8428 /time ), (DATASET 10954 /lat ),
(2): (DATASET 11377 /lon )
}
}
}
}
}
Step 2: Linking dimensions
In the second step another netCDF file “data.nc” is created. It has the same structure as the file in the previous step, but the dimensions are connected to the “grid.nc” file. Unlimited dimensions are not supported at the moment. They are converted to limited ones, as you can see in the output of ncdump.
Download: mklink.c
Output of ncdump:
$ ncdump -h data.nc
netcdf data {
dimensions:
lat = 6 ;
lon = 5 ;
time = 4 ;
variables:
float lat(lat) ;
float lon(lon) ;
float time(time) ;
int var1(time, lat, lon) ;
}
Output of h5dump:
$ h5dump -A -p data.nc
HDF5 "data.nc" {
GROUP "/" {
ATTRIBUTE "_NCProperties" {
DATATYPE H5T_STRING {
STRSIZE 8192;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_UTF8;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "version=1|netcdflibversion=4.4.1-rc2|hdf5libversion=1.10.0"
}
}
DATASET "lat" {
DATATYPE H5T_IEEE_F32LE
DATASPACE SIMPLE { ( 6 ) / ( 6 ) }
STORAGE_LAYOUT {
MAPPING 0 {
VIRTUAL {
SELECTION REGULAR_HYPERSLAB {
START (0)
STRIDE (1)
COUNT (1)
BLOCK (6)
}
}
SOURCE {
FILE "grid.nc"
DATASET "lat"
SELECTION REGULAR_HYPERSLAB {
START (0)
STRIDE (1)
COUNT (1)
BLOCK (6)
}
}
}
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE H5D_FILL_VALUE_DEFAULT
}
ATTRIBUTE "CLASS" {
DATATYPE H5T_STRING {
STRSIZE 16;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "DIMENSION_SCALE"
}
}
ATTRIBUTE "NAME" {
DATATYPE H5T_STRING {
STRSIZE 4;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "lat"
}
}
ATTRIBUTE "REFERENCE_LIST" {
DATATYPE H5T_COMPOUND {
H5T_REFERENCE { H5T_STD_REF_OBJECT } "dataset";
H5T_STD_I32LE "dimension";
}
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): {
DATASET 15866 /var1 ,
1
}
}
}
ATTRIBUTE "_Netcdf4Dimid" {
DATATYPE H5T_STD_I32LE
DATASPACE SCALAR
DATA {
(0): 0
}
}
}
DATASET "lon" {
DATATYPE H5T_IEEE_F32LE
DATASPACE SIMPLE { ( 5 ) / ( 5 ) }
STORAGE_LAYOUT {
MAPPING 0 {
VIRTUAL {
SELECTION REGULAR_HYPERSLAB {
START (0)
STRIDE (1)
COUNT (1)
BLOCK (5)
}
}
SOURCE {
FILE "grid.nc"
DATASET "lon"
SELECTION REGULAR_HYPERSLAB {
START (0)
STRIDE (1)
COUNT (1)
BLOCK (5)
}
}
}
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE H5D_FILL_VALUE_DEFAULT
}
ATTRIBUTE "CLASS" {
DATATYPE H5T_STRING {
STRSIZE 16;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "DIMENSION_SCALE"
}
}
ATTRIBUTE "NAME" {
DATATYPE H5T_STRING {
STRSIZE 4;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "lon"
}
}
ATTRIBUTE "REFERENCE_LIST" {
DATATYPE H5T_COMPOUND {
H5T_REFERENCE { H5T_STD_REF_OBJECT } "dataset";
H5T_STD_I32LE "dimension";
}
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): {
DATASET 15866 /var1 ,
2
}
}
}
ATTRIBUTE "_Netcdf4Dimid" {
DATATYPE H5T_STD_I32LE
DATASPACE SCALAR
DATA {
(0): 1
}
}
}
DATASET "time" {
DATATYPE H5T_IEEE_F32LE
DATASPACE SIMPLE { ( 4 ) / ( H5S_UNLIMITED ) }
STORAGE_LAYOUT {
CHUNKED ( 1024 )
SIZE 4096
}
FILTERS {
NONE
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE 9.96921e+36
}
ALLOCATION_TIME {
H5D_ALLOC_TIME_INCR
}
ATTRIBUTE "CLASS" {
DATATYPE H5T_STRING {
STRSIZE 16;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "DIMENSION_SCALE"
}
}
ATTRIBUTE "NAME" {
DATATYPE H5T_STRING {
STRSIZE 5;
STRPAD H5T_STR_NULLTERM;
CSET H5T_CSET_ASCII;
CTYPE H5T_C_S1;
}
DATASPACE SCALAR
DATA {
(0): "time"
}
}
ATTRIBUTE "REFERENCE_LIST" {
DATATYPE H5T_COMPOUND {
H5T_REFERENCE { H5T_STD_REF_OBJECT } "dataset";
H5T_STD_I32LE "dimension";
}
DATASPACE SIMPLE { ( 1 ) / ( 1 ) }
DATA {
(0): {
DATASET 15866 /var1 ,
0
}
}
}
ATTRIBUTE "_Netcdf4Dimid" {
DATATYPE H5T_STD_I32LE
DATASPACE SCALAR
DATA {
(0): 2
}
}
}
DATASET "var1" {
DATATYPE H5T_STD_I32LE
DATASPACE SIMPLE { ( 4, 6, 5 ) / ( H5S_UNLIMITED, 6, 5 ) }
STORAGE_LAYOUT {
CHUNKED ( 1, 6, 5 )
SIZE 480
}
FILTERS {
NONE
}
FILLVALUE {
FILL_TIME H5D_FILL_TIME_IFSET
VALUE -2147483647
}
ALLOCATION_TIME {
H5D_ALLOC_TIME_INCR
}
ATTRIBUTE "DIMENSION_LIST" {
DATATYPE H5T_VLEN { H5T_REFERENCE { H5T_STD_REF_OBJECT }}
DATASPACE SIMPLE { ( 3 ) / ( 3 ) }
DATA {
(0): (DATASET 9250 /time ), (DATASET 8428 /lat ),
(2): (DATASET 8842 /lon )
}
}
}
}
}
Future Work
In the next step we plan to integrate the external dimensions in our workflows.